Material below is adapted from the SfN Short Course Distinct Molecular Programs Define Human Radial Glia Subtypes During Human Cortical Development, by Tomasz J. Nowakowski, PhD, and Alex A. Pollen, PhD. Short Courses are day-long scientific trainings on emerging neuroscience topics and research techniques held the day before SfN’s annual meeting.
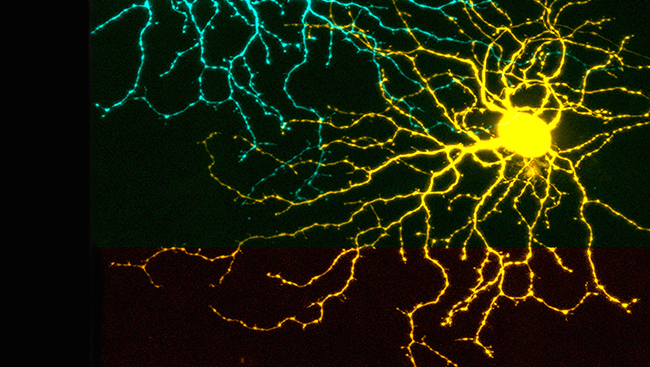
Multicellular organisms were able to evolve in part because of the capability of cells to take specialized shapes and perform varied functions. Understanding how cells diversify their characteristics has become simpler with the rise of single-cell RNA sequencing, which allows scientists to learn more about the genes and pathways active in an individual cell. Researchers can apply this technique to learn more about development of the human nervous system.
Access to the full article is available to SfN members.
Neuronline is a benefit of SfN membership. Renew your membership now to make sure you don’t lose access.