Material below is adapted from the SfN Short Course Modeling Predisposition to Schizophrenia, a Genetically Heterogeneous Neuropsychiatric Disorder, Using Induced Pluripotent Stem Cells, by Seok-Man Ho, BSc, Erin Flaherty, BSc, and Kristen J. Brennand, PhD. Short Courses are day-long scientific trainings on emerging neuroscience topics and research techniques held the day before SfN’s annual meeting.
Schizophrenia is a chronic and severe mental disorder affecting approximately one percent of the world population. In recent years, scientists have employed animal models and genome-wide association studies to gain important insights into the biological underpinnings of schizophrenia. However, it remains unclear exactly how environmental and genetic factors interact to increase the risk for this debilitating disease.
Human induced pluripotent stem cells (iPSCs) have the potential to shed new light on this question. Using skin or blood cells from individual patients and genetically reprogramming the to revert to an embryonic stem cell-like state, scientists can then convert these cells into mature cell types retaining the identical genetic makeup of the donor individual. By focusing on iPSCs drawn from patients with common clinical manifestations or rare genetic variants, researchers hope to unravel the complex genetics of this disease.
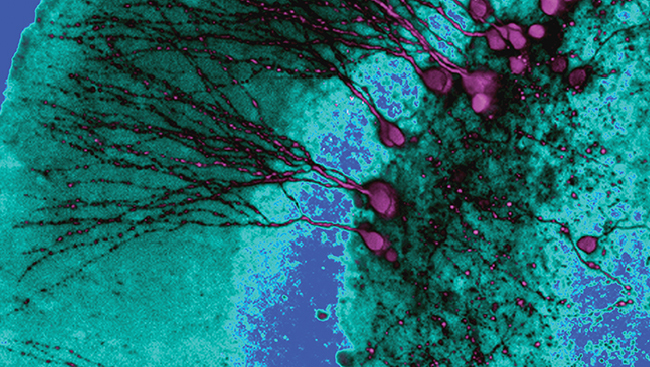
Modeling neuropsychiatric disease requires researchers to generate different types of neurons. They do that either through directed differentiation or neuronal induction. Both techniques can generate functional, disease-relevant cell types, including dopaminergic, glutamatergic, and GABAergic neurons.
Directed differentiation is an in vitro strategy recapitulating in vivo development by exposing iPSCs to small molecules or morphogens, which mimic the signaling involved in the patterning, specification, and commitment of defined cell types during embryonic development. Scientists can generate highly pure populations of neurons by treating iPSCs with molecules that inhibit bone morphogenetic protein and transforming growth factor-beta — a process known as dual SMAD inhibition.
However, one major drawback of directed differentiation is the nearly three months needed to generate functional neurons. Recently, researchers established neuronal induction as a viable alternative strategy to rapidly and directly convert patient-derived somatic cells into a variety of neurons. Forced expression of key transcription factors is sufficient to change cell fate from one committed cell type to another within just a few weeks.
Regardless of which method is used to generate neurons, important concerns remain regarding the utility of iPSCs for disease modeling. For example, the relatively small sample size of human iPSC-based studies makes it difficult to fully capture the heterogeneity among patients with schizophrenia. To overcome this challenge, researchers can select patient cohorts based on two different criteria: a shared genetic mutation or a similar clinical profile. The first strategy parallels traditional mouse-based studies that investigate the effects of rare loci, while the second approach takes full advantage of the ability of human iPSC-based studies to investigate complex genetic disorders without full knowledge of all the genes involved.
Much more work needs to be done to fully harness the potential of iPSCs for neuropsychiatric disease modeling. One major challenge will be creating more complex neural circuits that better resemble those in the human brain. To this end, iPSC-based models should incorporate multiple neuronal cell types together with glial cells, as well as stimulation with hormones and other environmental factors that promote synaptic plasticity. Alternatively, each of these human cell types could be transplanted into mouse models of disease, allowing these cells to integrate into existing neural circuits in the mouse brain.
To uncover the common pathways disrupted in schizophrenia, researchers will need to combine data from iPSC-based studies, genome-wide association studies, and mouse models. Moreover, experiments must gather multilayered information about the patient’s clinical, genomic, transcriptomic, proteomic, and epigenomic profiles. Together, this rich set of information could reveal the key drivers of complex genetic disorders to guide the development of new therapeutics.