Material below summarizes the article A New Mouse Line Reporting the Translation of Brain-Derived Neurotrophic Factor Using Green Fluorescent Protein, published on December 27, 2019, in eNeuro and authored by Erin Wosnitzka, Xinsheng Nan, Jeff Nan, Pedro Chacón-Fernández, Lothar Kussmaul, Michael Schuler, Bastian Hengerer, and Yves-Alain Barde.
Highlights
- The direct coupling of the secretory protein brain-derived neurotrophic factor (BDNF) with green fluorescent protein (GFP) interferes with its biosynthesis and secretion.
- Translation can nonetheless be monitored using constructs translating BDNF and GFP as separate proteins.
- To avoid the artefacts resulting from cDNA overexpression, the strategy also involves a substitution of the genomic protein coding sequence, as previously reported with the cytoplasmic protein BRAF.
 |
|
|
Study Question
Can GFP fusion constructs be used to explore the physiology and cell biology of BDNF, as much of the current literature suggests? If not, is there an alternative that would also make drug screening possible in a brain-region-specific manner?
How This Research Advances What We Know
Like many growth factors, BDNF acts at very low concentrations through the activation of high affinity receptors. While the restricted availability and low abundance of growth factors is a key aspect of their specificity, these attributes also make the detection of growth factors in cells and tissues exceedingly difficult.
Furthermore, the lack of thoroughly validated antibodies specifically recognizing their targets continues to be a major problem. The first BDNF monoclonal antibody suitable for Western blotting only became available recently, and there is just one fully validated monoclonal antibody allowing the detection of BDNF by immunohistochemistry. These difficulties explain why over a long period of time, scientists resorted to overexpression paradigms under the assumption that the distribution of overexpressed BDNF may reflect the distribution of endogenous BDNF.
Experimental Design or Methodology
The study had three main components:
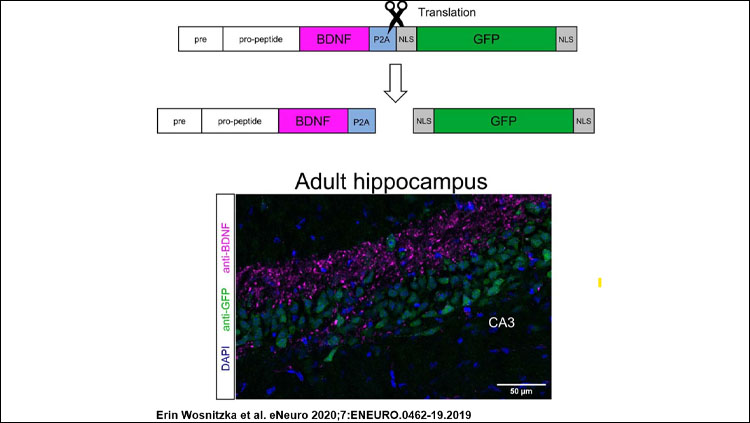
- Use of expression plasmids encoding BDNF tagged with GFP, with GFP preceded by a virus-derived P2A self-cleaving sequence or with up to four 10-amino acid Myc epitopes. The processing of BDNF is examined by Western blot using a monoclonal antibody validated using brain tissue from animals lacking Bdnf. The conditioned media collected from the transfected cells is also used to quantify the activation of the BDNF tyrosine kinase receptor TrkB.
- Localization of endogenous BDNF in cultured neurons as well as BDNF and GFP within neurons isolated from animals carrying a Bdnf gene substitution (Bdnf replaced by Bdnf-P2a-Gfp), and comparison with the results obtained from neurons transfected with BDNF expression plasmids.
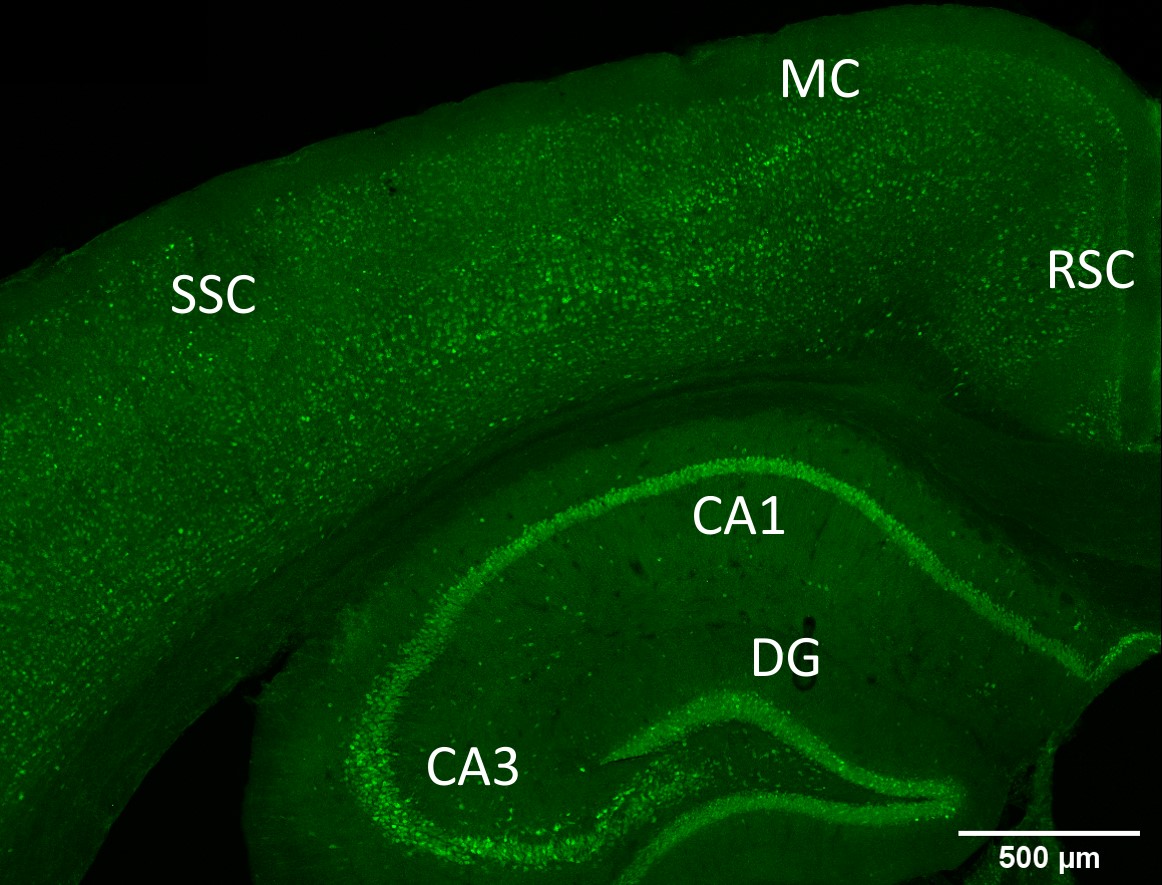
- Localization of BDNF and GFP on brain sections prepared from the BDNF-P2A-GFP animals and quantification of the GFP signal in the hippocampus of these animals.
Results
The direct coupling of BDNF to GFP turns out to impair the processing of the BDNF precursor as well as the secretion of BDNF. The study also illustrates the observation that tags of increasing length proportionally compromise the ability of BDNF to fully activate TrkB.
This is proposed to be the explanation for the mild obesity phenotype observed in ageing animals carrying the Bdnf-P2a-Gfp gene substitution: The 22-amino acid P2A sequence remaining attached to BDNF is likely to chronically decrease the activation of TrkB. This suggestion is based on abundant literature, including human genetics, indicating that a fully functional BDNF/TrkB pathway decreases food intake.
Most importantly, BDNF translation can be assessed in brain sections by monitoring the intensity of the GFP signal. The quantitative data indicate a near-perfect match with previous in situ hybridization studies suggesting that under physiological conditions, BDNF protein levels are primarily regulated by transcriptional activity. Curiously, these results do not quite match published data obtained by single cell RNAseq for reasons that remain unclear.
Interpretation
The widely used approach of transfecting cells with cDNA constructs to explore protein localization and/or trafficking should critically examine the possibility that the distribution of the over-expressed protein may not match the distribution of the endogenous protein. This is not the case for BDNF.
Long tags such as GFP would seem a priori problematic when fused with proteins undergoing extensive co- and posttranslational processing including glycosylation, homodimerization, proteolytic cleavage, and vesicular packaging and trafficking.
Reliable visualization of the trafficking of endogenous proteins will require new developments including much smaller genetically encoded probes.
With the approach described in our contribution, single cells can now be isolated from mouse tissues and their molecular make-up correlated with the degree of BDNF translation.
The project was the result of a close collaboration between groups in Cardiff and Biberach: Erin Wosnitzka, Xinsheng Nan, and Yves Barde, at Cardiff University, Jeff Nan and Pedro Chacon-Fernandez, who used to work at Cardiff University, and Lothar Kussmaul, Michael Schuler, and Bastian Hengerer, at Boehringer Ingelheim, a pharmaceutical company in Germany.
Visit eNeuro to read the original article and explore other content. Read other summaries of eNeuro and JNeurosci papers in the Neuronline collection SfN Journals: Research Article Summaries.
A New Mouse Line Reporting the Translation of Brain-Derived Neurotrophic Factor Using Green Fluorescent Protein. Erin Wosnitzka, Xinsheng Nan, Jeff Nan, Pedro Chacón-Fernández, Lothar Kussmaul, Michael Schuler, Bastian Hengerer, and Yves-Alain Barde. eNeuro 27 December 2019, 7 (1) ENEURO.0462-19.2019; DOI: 10.1523/ENEURO.0462-19.2019.